Welcome to MEDCIS Cross Cohort Query Interface
- Query Builder, with terminology support and visual controls such as slider bar and checkboxes;
- Query Manager, which stores and labels queries for reuse.
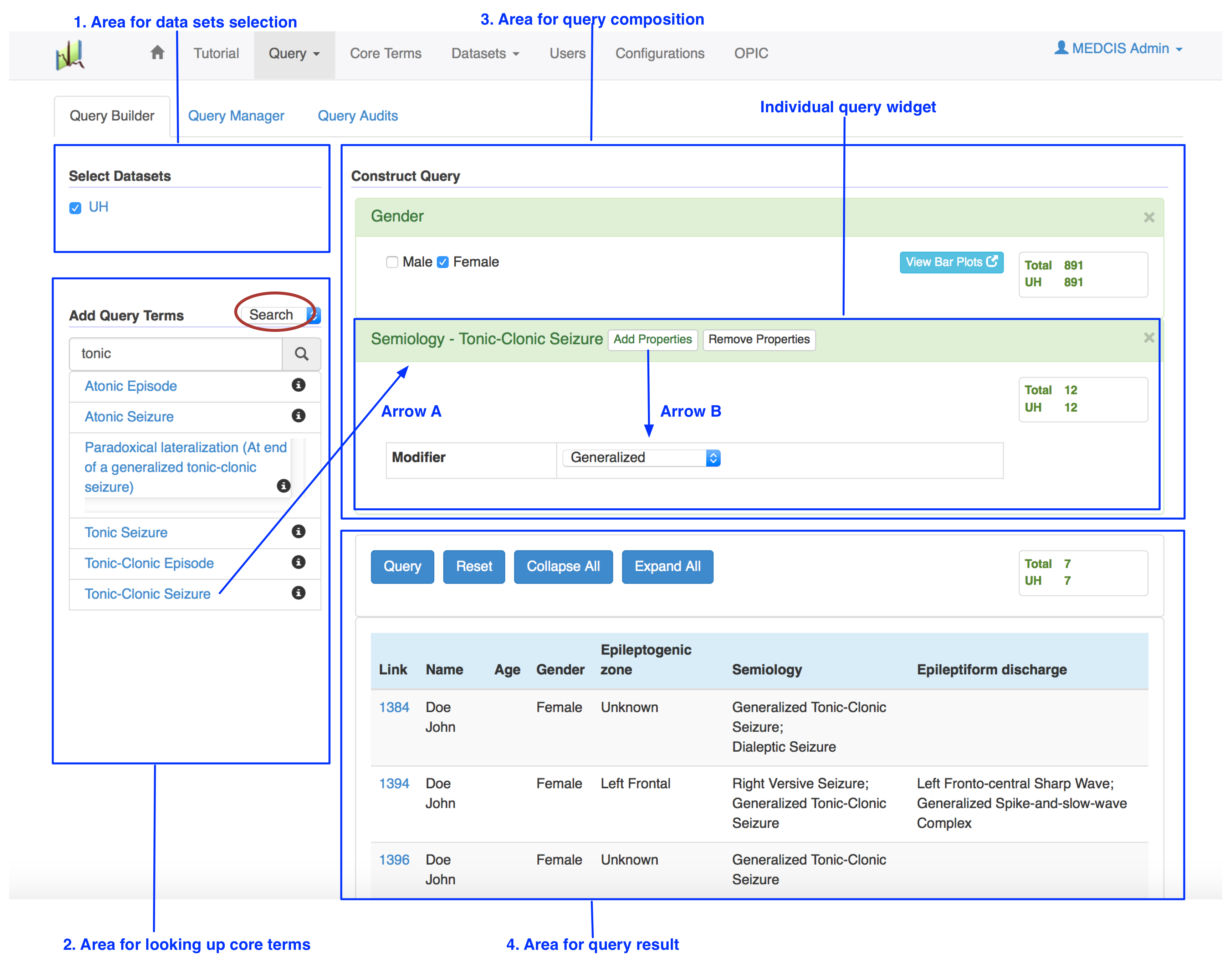
Clicking the sub menu Query Builder under the menu Query will lead you to the Query Builder page[Fig01].
- In the "Area for data sets selection" (blue box 1), you can select datasets of interest such as "UH".
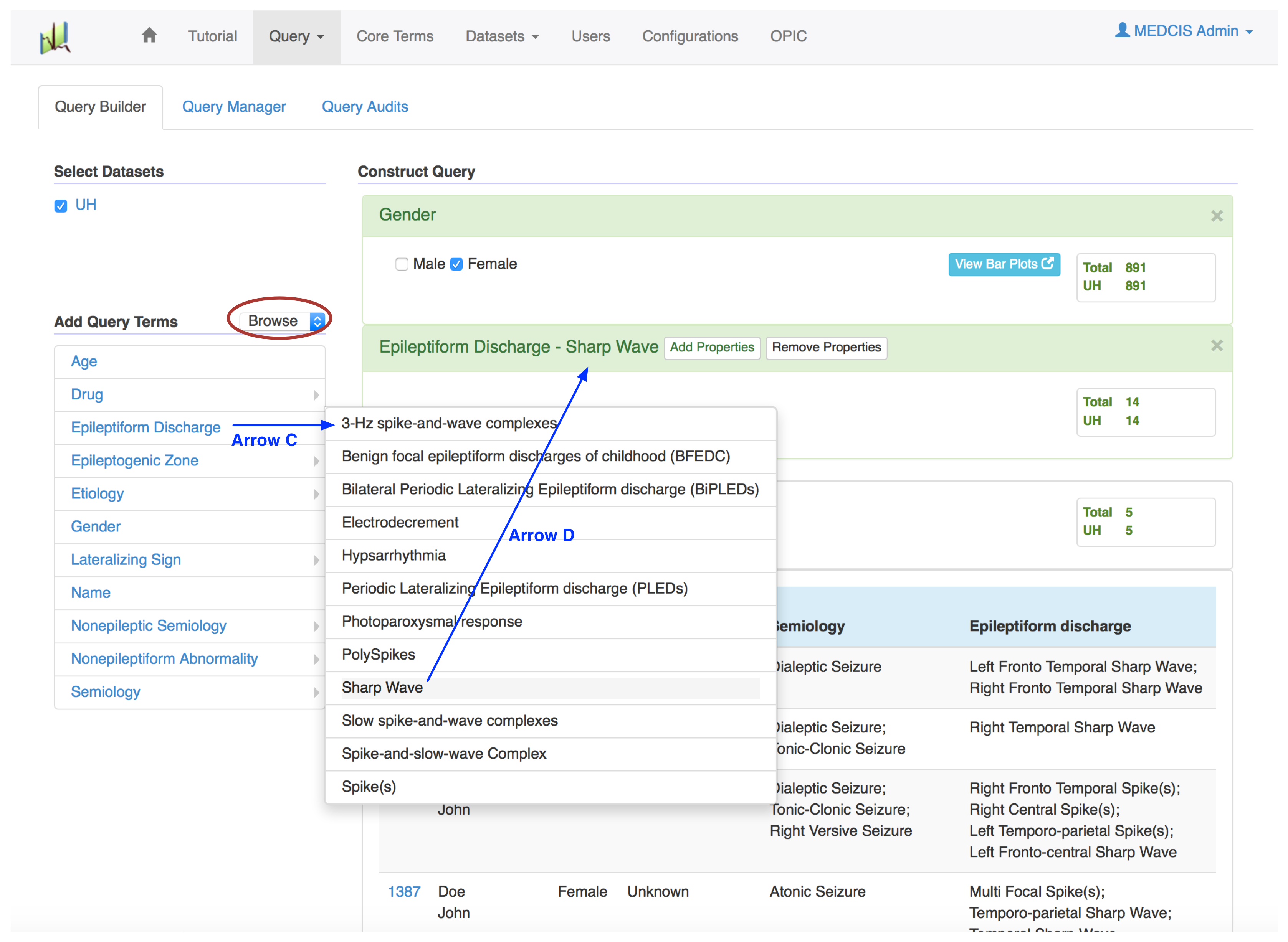
- In the "Area for looking up core terms" (blue box 2), you can look up a specific core term in two ways. The first way is "search" (red circle), where you can search for core terms like "tonic" in the text box. Then a list of candidate variable terms, which partially match the search term, will be automatically displayed. The second way is "browse" (red circle in Fig02), where you can browse core terms in terms of categories. For example, clicking the category "Epileptiform Discharge" (Arrow C) will pop up a sub menu with all the core terms or sub categories under this category.
- Clicking an appropriate candidate term (Arrow A) will add a corresponding individual query widget to the "Area for query composition" (blue box 3), where you can specify the query based on the type of the term. If the term is a categorical type, then checkboxes will be provided for you to select. If the term is a numeric type, then a slider bar along with two text boxes specifying range will be provided so that you can either slide the bar or enter a range in the text boxes. If the term is a hierarchical type, then a list of all subtypes of this term will be provided for you to select or further drill down to more specific level.
- Clicking the button "Add Properties" (Arrow B) in a query widget will display all the properties of the core term for you to further specify query criteria (e.g., choosing "Generalized" for the property "Modifier").
- Clicking the "Query" button on the top left corner of the "Area for query result" (blue box 4) will generate the patient reports satisfying the composed query criteria. For example, Fig01 shows a sample query for UH patient reports where patients are female and with generalized tonic-clonic seizure. Fig02 shows a sample query for UH patient reports where patients are female and with epileptiform discharge sharp wave.
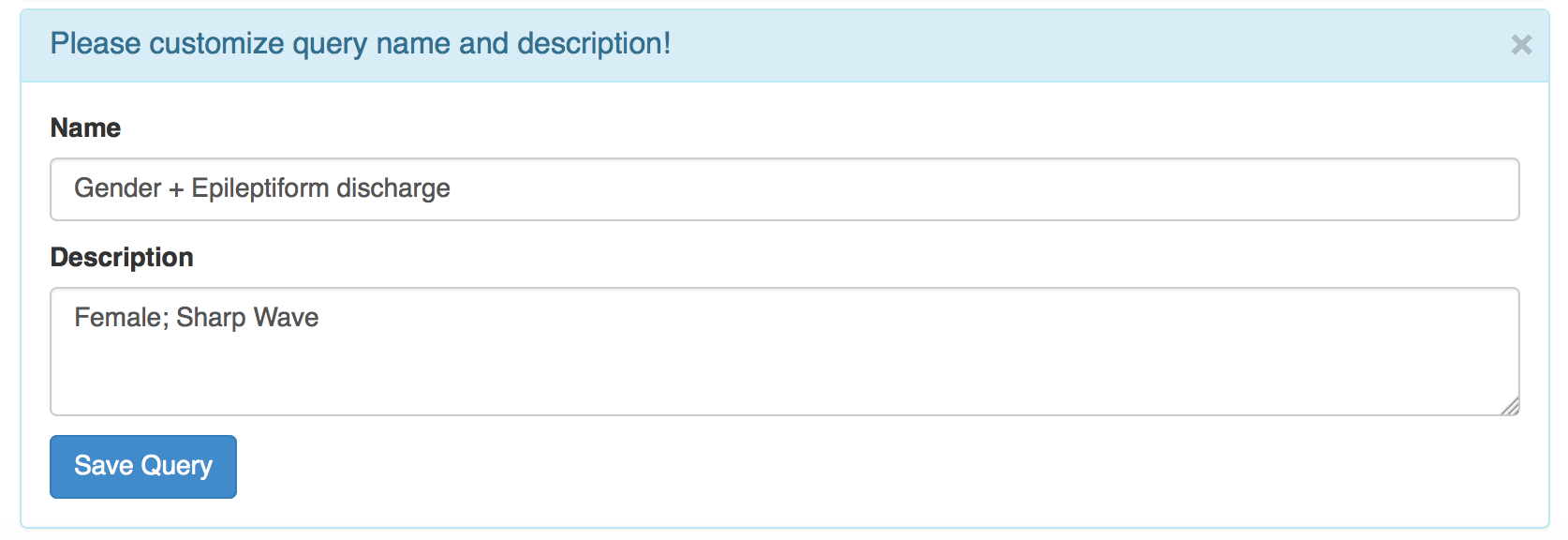
While you are exploring or done with the query composition, you can also scroll down and save the constructed query[Fig03].
2. Query Manager
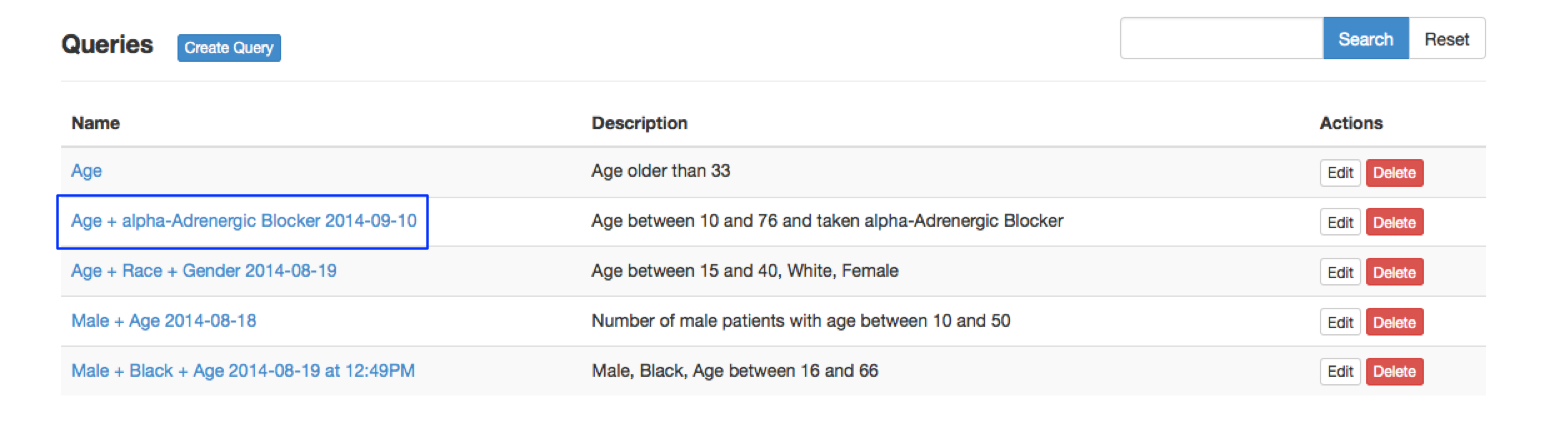
Clicking the "Save Query" button in Fig03 will lead you to the Query Manager page[Fig04], which allows you to keep track of saved queries and retrieve record information on the queries. You can also click the sub menu Query Manager under the menu Query or click the tab Query Manager to get to the Query Manager page[Fig04]. Clicking a name of a saved query (blue box) will link to the Query Builder page with query widgets automatically loaded for the saved query, where you can review and update query criteria.
Ready to build your own query? Build Query
Download for Windows
MEDCIS uses Ruby language and Command Prompt for batch data downloads. Here's the guide for running Ruby in the Windows.
1. Install Ruby environment
You can install Ruby using Ruby installer. Go to their website and get the latest version.https://rubyinstaller.org/
Once you install Ruby, click the Windows Start Button, search CMD, and open the Command Prompt program.
You can check if the Ruby is installed by typing:
2. Install CSR Data Inventory Gem
The CSR Data Inventory Gem is a package we developed to assist your downloading. Open the Command Prompt and typing:3. Request Data Download Privilege
Please sign up an MEDCIS account first and send your personal and project information to . We will grant the download permission to your account.
4. Get Download Token for a Dataset
The download token is the unique code for each download request. You will be asked to enter it when start the download.
Download Token of a whole siteYou can generate token to download the whole site. copy the token and see step 5.
Download Token of your customized query
You can generate token for your customized query. To do so, use the query builder and get the result, and click the download button to save the query and generate the token. You can also select the data types to narrow down the scope of your download.
The saved query can be found on the Query Manager.
5. Download Data with Ruby
Go back to your Command Prompt and type in the lines below:
1. create new folder for downloadcd your_folder_name
You can see the download process begins. It might take some time depend on amount of files.
5. FAQs
How do I update the query for download?Repeat step 3 and 4 if the dataset need to be updated.
Download for Mac/Linux
MEDCIS uses Ruby language for batch data downloads. Here's the guide for Mac/Linux.
1. Install Ruby environment
Your Mac and Linux might already have Ruby installed by default. You can check with the Terminal by typing:
If there's a version of Ruby, you can move on to step 2.
If you don't have a version of Ruby, please follow the Ruby official document to install.
2. Install CSR Data Inventory Gem
The CSR Data Inventory Gem is a package we developed to assist your downloading. Open the Terminal and typing:3. Request Data Download Privilege
Please sign up an MEDCIS account first and send your personal and project information to . We will grant the download permission to your account.
4. Get Download Token for a Dataset
The download token is the unique code for each download request. You will be asked to enter it when start the download.
Download Token of a whole siteYou can generate token to download the whole site. copy the token and see step 5.
Download Token of your customized query
You can generate token for your customized query. To do so, use the query builder and get the result, and click the download button to save the query and generate the token. You can also select the data types to narrow down the scope of your download.
The saved query can be found on the Query Manager.
t
Go back to your terminal and type the lines below:
1. create new folder for downloadcd your_folder_name
CSRDataInventory.download;
You can see the download process begins. It might take some time depending on the amount of files.
5. FAQs
How do I update the query for download?Repeat step 3 and 4 if the dataset need to be updated.